1 Abstract¶
A catalog of bright stars with clean surrounding is required to perform focus and alignment with the AuxTel. It is anticipated that rebuilding star catalog is required when new criteria should be applied to the selection of stars. This technote is to describe the notebook that creates a new catalog based on queries from Tycho-2 and HD catalog. This notebook can be found on this github repo https://github.com/lsst-sitcom/sitcomtn-058/tree/main/_static. Please note that it is recommended to run this notebook in a local environment as it may take several tens of minutes or longer to complete.
2 Generating a star catalog for the Auxtel observations¶
The following description is about the Jupyter notebook used to generate the star catalog with the required selection criteria. See also details about focusing, center, and absorb pointing offsets at AuxTel.
2.1 Create a notebook with the Tycho-2 catalog¶
First, a list of stars from Vizier query of
Tycho-2 catalogue [2].
The default selection criteria is all stars with DEmdeg = '<10.0' and VTmag = '<10' and the name of output file is file = './cwfs_tycho2_stars.pd'.
The total number of the stars with Decl. < 10 deg, Vmag < 10mag from Tycho-2 catalog is 193,650.
2.2 Cutting Selection Criteria¶
If this is your first time to run this Jupyter notebook, the Tycho-2 catalog will be queried by constraints from Vizier. Then, the list of all stars matching the constraints will be saved into a JSON file file = './cwfs_tycho2_stars.pd'.
If the file exists locally, the list of all stars will be directly read from the JSON file without querying.
if os.path.exists(file):
print('Read existing Vizier Catalog in Local'+file)
tycho_star_all = pd.read_json(file)
else :
print('Read Vizier Catalog I/259/tyc2 ')
tycho_star_all = Vizier.query_constraints(catalog='I/259/tyc2', DEmdeg=DEmdeg, VTmag=VTmag)[0]
tycho_star_all = tycho_star_all.to_pandas()
result = tycho_star_all.to_json(file)
In order to exclude targets contaminated by bright nearby stars, :
Include stars with 6 < V < 8
Exclude stars with
bright (V < 10 mag) nearby stars within 1 arcmin. radius
very bright (V < 6 mag) stars within 7 arcmin radius
extremely bright (V < 3 mag) stars within 20 arcmin radius
Exclude stars with higher proper motion (> 100 mas/yr)
mag_upper = 6 ; mag_lower = 8; pm_cut = 100 #pm_cut for net pm (sqrt(pmRA*cosDec^2.0+pmDEC^2.0))
area_cut1 = 1; area_cut2 = 7; area_cut3 = 20 #arcmin.
mag_cut1 = 10; mag_cut2 = 6; mag_cut3= 3 #vmag
tycho_star_all['net_pm'] = (((tycho_star_all['pmRA'])**2.0 + (tycho_star_all['pmDE'])**2.0)**0.5)
tycho_star_cut = tycho_star_all[(tycho_star_all['VTmag'] > mag_upper) & (tycho_star_all['VTmag'] < mag_lower) & (tycho_star_all['net_pm'] < pm_cut)]
def sep(RA1,RA2,Dec1,Dec2):
sep = ((((RA1-RA2)*np.cos(Dec1))**(2.0))+((Dec1-Dec2)**2.0))**(0.5)
return sep
tycho_star_cut_criteria = pd.DataFrame.copy(tycho_star_cut)
for i in tycho_star_cut.index:
within_1arcmin = (sep(tycho_star_cut["RA_ICRS_"][i],tycho_star_all["RA_ICRS_"],tycho_star_cut["DE_ICRS_"][i],tycho_star_all["DE_ICRS_"]) < area_cut1/60.0) & (tycho_star_all["VTmag"] < mag_cut1)
within_7arcmin = (sep(tycho_star_cut["RA_ICRS_"][i],tycho_star_all["RA_ICRS_"],tycho_star_cut["DE_ICRS_"][i],tycho_star_all["DE_ICRS_"]) < area_cut2/60.0) & (tycho_star_all["VTmag"] < mag_cut2)
within_20arcmin = (sep(tycho_star_cut["RA_ICRS_"][i],tycho_star_all["RA_ICRS_"],tycho_star_cut["DE_ICRS_"][i],tycho_star_all["DE_ICRS_"]) < area_cut3/60.0)& (tycho_star_all["VTmag"] < mag_cut3)
if np.count_nonzero(within_1arcmin) > 1 or np.count_nonzero(within_7arcmin) > 0 or np.count_nonzero(within_20arcmin) > 0:
tycho_star_cut_criteria.drop([i], inplace=True)
TYC1 |
TYC2 |
TYC3 |
pmRA |
pmDE |
BTmag |
VTmag |
HIP |
RA_ICRS |
DE_ICRS |
net_pm |
|
|---|---|---|---|---|---|---|---|---|---|---|---|
1 |
4663 |
45 |
1 |
45.90000 |
-53.400002 |
7.695 |
6.420 |
417 |
1.265827 |
-0.502912 |
35.208 |
6 |
4663 |
160 |
1 |
62.20000 |
-15.300000 |
8.819 |
7.399 |
14 |
0.048280 |
-0.360421 |
32.027 |
7 |
4663 |
363 |
1 |
7.600000 |
-4.200000 |
7.972 |
6.349 |
664 |
2.050386 |
-2.447699 |
4.3417 |
25 |
4663 |
1285 |
1 |
40.20000 |
-4.700000 |
8.318 |
7.164 |
700 |
2.179543 |
-2.222573 |
20.237 |
36 |
4664 |
560 |
1 |
-9.100000 |
35.400002 |
8.027 |
7.051 |
1587 |
4.961996 |
-2.478966 |
18.275 |
… |
… |
… |
… |
… |
… |
… |
… |
… |
… |
… |
… |
193594 |
1170 |
1 |
1 |
-49.5000 |
-182.20000 |
8.179 |
7.538 |
117236 |
356.5766 |
9.785502 |
94.402 |
193598 |
1170 |
247 |
1 |
16.79999 |
-21.10000 |
9.190 |
7.819 |
116978 |
355.7045 |
9.885516 |
13.486 |
193605 |
1170 |
883 |
1 |
31.00000 |
-26.50000 |
7.024 |
6.684 |
117394 |
357.0505 |
8.245658 |
20.391 |
193626 |
1171 |
425 |
1 |
15.60000 |
-8.800000 |
6.827 |
6.818 |
117962 |
358.9071 |
8.223298 |
8.955 |
193636 |
1171 |
1243 |
1 |
10.20000 |
0.3000000 |
9.586 |
7.936 |
117912 |
358.7502 |
8.389472 |
5.102 |
2.3 Match Tycho-2 and HD catalogs¶
Now the star list is matched to HD identifications for Tycho-2 stars [1]
Query Vizier Catalog IV/25/tyc2.
print('Query Vizier Catalog IV/25/tyc2_hd to match TYC ID and HD ID')
HD_stars_all = Vizier.query_constraints(catalog='IV/25/tyc2_hd')[0]
Add
pmRA,pmDE,net_pmfields.
index_only = HD_stars_all[0:0]
index_only.add_columns([(),(),()],names=['pmRA','pmDE','net_pm'])
index_only['pmRA'] = index_only['pmRA'].astype(np.float32)
index_only['pmDE'] = index_only['pmDE'].astype(np.float32)
index_only['net_pm'] = index_only['net_pm'].astype(np.float32)
HD_star_match= Table(index_only)
Match Tycho-2 and HD catalogs.
for i in tycho_star_cut_criteria.index:
condition = (HD_stars_all["TYC1"]==tycho_star_cut_criteria["TYC1"][i]) & \
(HD_stars_all["TYC2"]==tycho_star_cut_criteria['TYC2'][i]) & \
(HD_stars_all["TYC3"]==tycho_star_cut_criteria['TYC3'][i])
if np.count_nonzero(condition) == 1:
table = HD_stars_all[condition]
table['pmRA'] = tycho_star_cut_criteria["pmRA"][i]
table['pmDE'] = tycho_star_cut_criteria["pmDE"][i]
table['net_pm'] = tycho_star_cut_criteria["net_pm"][i]
HD_star_match.add_row(table[:][0])
2.4 Query Simbad data for star sample¶
Subsequently, a query of the Simbad database is executed for each of the selected CWFS stars.
The default VOTable fields:
MAIN_ID,RA,DEC,RA_PREC,DEC_PREC,COO_ERR_MAJA,COO_ERR_MINA,COO_ERR_ANGLE,COO_QUAL,COO_WAVELENGTH,COO_BIBCODE,SCRIPT_NUMBER_IDAdd
flux_name(V),flux(V),flux_error(V),flux_system(V),flux_bibcode(V),flux_qual(V),flux_univ(V)VOTable fields.Add
HD_ID,pmRA,pmDE,net_pmfields on the tabale.If you need another Simbad VOTable fields, check
Simbad.list_votable_fields()and add fields usingadd_votable_fields().
customSimbad = Simbad()
customSimbad.add_votable_fields('flux_name(V)', 'flux(V)', 'flux_error(V)', 'flux_system(V)','flux_bibcode(V)', 'flux_qual(V)', 'flux_unit(V)')
for i in range(0,len(HD_star_match["HD"])):
result_table = customSimbad.query_object('HD '+str(HD_star_match["HD"][i]))
result_table["HD_ID"] = 'HD '+str(HD_star_match["HD"][i])
result_table["pmRA"] = HD_star_match["pmRA"][i]
result_table["pmDE"] = HD_star_match["pmDE"][i]
result_table["net_pm"] = HD_star_match["net_pm"][i]
if i==0:
final = result_table
else:
final.add_row(result_table[:][0])
MAIN_ID |
RA |
DEC |
RA_PREC |
DEC_PREC |
COO_ERR_MAJA |
COO_ERR_MINA |
COO_ERR_ANGLE |
COO_QUAL |
COO_WAVELENGTH |
COO_BIBCODE |
FILTER_NAME_V |
FLUX_V |
FLUX_ERROR_V |
FLUX_SYSTEM_V |
FLUX_BIBCODE_V |
FLUX_QUAL_V |
FLUX_UNIT_V |
SCRIPT_NUMBER_ID |
HD_ID |
pmRA |
pmDE |
net_pm |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
HD 6 |
00 05 03.8227 |
-00 30 10.928 |
14 |
14 |
0.037 |
0.023 |
90 |
A |
O |
2020yCat.1350….0G |
V |
6.298 |
0.010 |
Vega |
2000A&A…355L..27H |
D |
V |
1 |
HD 6 |
45.9 |
-53.4 |
70.416 |
HD 224726 |
00 00 11.6217 |
-00 21 37.608 |
14 |
14 |
0.021 |
0.017 |
90 |
A |
O |
2020yCat.1350….0G |
V |
7.27 |
– |
Vega |
E |
V |
1 |
HD 224726 |
62.2 |
-15.3 |
64.054 |
|
* 5 Cet |
00 08 12.0955 |
-02 26 51.740 |
14 |
14 |
0.059 |
0.036 |
90 |
A |
O |
2020yCat.1350….0G |
V |
6.22 |
– |
Vega |
E |
V |
1 |
HD 352 |
7.6 |
-4.2 |
8.683 |
|
HD 406 |
00 08 43.1091 |
-02 13 21.296 |
14 |
14 |
0.036 |
0.030 |
90 |
A |
O |
2020yCat.1350….0G |
V |
7.05 |
0.010 |
Vega |
2000A&A…355L..27H |
D |
V |
1 |
HD 406 |
40.2 |
-4.7 |
40.474 |
HD 1567 |
00 19 50.8746 |
-02 28 43.990 |
14 |
14 |
0.028 |
0.015 |
90 |
A |
O |
2020yCat.1350….0G |
V |
6.96 |
0.010 |
Vega |
2000A&A…355L..27H |
D |
V |
1 |
HD 1567 |
-9.1 |
35.4 |
36.551 |
… |
… |
… |
… |
… |
… |
… |
… |
… |
… |
… |
… |
… |
… |
… |
… |
… |
… |
… |
… |
… |
… |
… |
HD 1421 |
00 18 18.5194 |
-02 00 53.291 |
14 |
14 |
0.021 |
0.014 |
90 |
A |
O |
2020yCat.1350….0G |
V |
7.18 |
– |
Vega |
E |
V |
1 |
HD 1421 |
35.2 |
-2.2 |
35.269 |
|
HD 999 |
00 14 24.4641 |
-02 11 52.802 |
14 |
14 |
0.022 |
0.018 |
90 |
A |
O |
2020yCat.1350….0G |
V |
7.18 |
0.010 |
Vega |
2000A&A…355L..27H |
D |
V |
1 |
HD 999 |
-11.1 |
-29.1 |
31.145 |
HD 820 |
00 12 40.3372 |
-01 13 37.885 |
14 |
14 |
0.019 |
0.015 |
90 |
A |
O |
2020yCat.1350….0G |
V |
7.2 |
0.010 |
Vega |
2000A&A…355L..27H |
D |
V |
1 |
HD 820 |
78.4 |
-0.2 |
78.400 |
HD 1369 |
00 17 48.3722 |
-01 51 45.858 |
14 |
14 |
0.068 |
0.055 |
90 |
A |
O |
2020yCat.1350….0G |
V |
7.1 |
0.010 |
Vega |
2000A&A…355L..27H |
D |
V |
1 |
HD 1369 |
-3.7 |
4.3 |
5.6727 |
HD 2023 |
00 24 29.6495 |
-02 13 08.626 |
14 |
14 |
0.027 |
0.018 |
90 |
A |
O |
2020yCat.1350….0G |
V |
6.067 |
0.010 |
Vega |
2000A&A…355L..27H |
D |
V |
1 |
HD 2023 |
-34.8 |
-42.4 |
54.853 |
2.5 Exclude individual stars manually (optional)¶
This section is to exclude the stars from the list manually.
Put the names of stars (format:HDNNNNNN) on the Remove_main_id parameter.
Remove_main_id = ["HD22746","HD452"] #HD NNNNNN
p= re.compile("\d*\.?\d+")
customSimbad = Simbad()
for i in range(len(Remove_main_id)):
number = p.findall(Remove_main_id[i])[0]
Remove_main_id_simbad= customSimbad.query_region('HD '+str(number))["MAIN_ID"]
Remove = (final["MAIN_ID"] == Remove_main_id_simbad[0])
if np.count_nonzero(Remove) !=0 :
remove_index = [i for i, x in enumerate(Remove) if x]
final.remove_row(remove_index[0])
print(Remove_main_id_simbad[0]+' is now removed from the final catalog')
2.6 Save the catalog on the output file¶
As a final step, the queried table is saved into json file.
The default name for output is HD_cwfs_stars.pd.
The file name can changed with file_name_final variable.
file_name_final = 'HD_cwfs_stars.pd' #file_name
result = final.to_pandas().to_json(file_name_final)
print('List of Stars was exported to '+file_name_final)
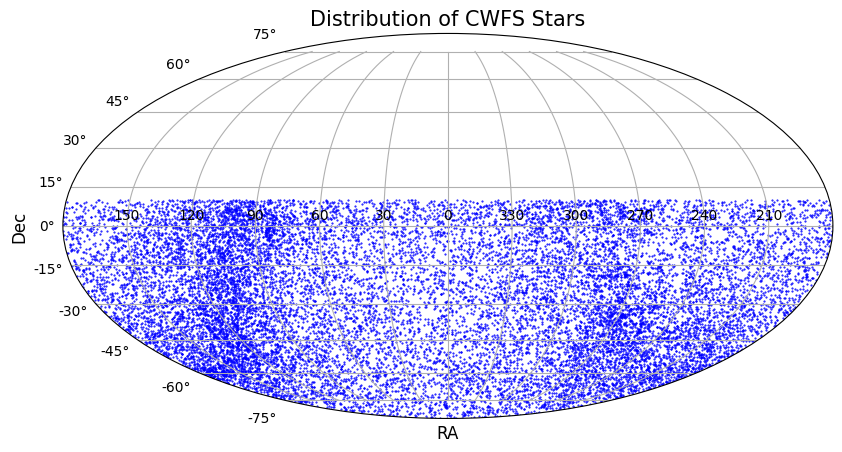
3 Plot for the distribution of the Stars on Sky¶
To check whether selected CWFS stars are homogeneously distributed on the southern sphere, equatorial coordinates RA, Dec of each starsare plotted on a Mollweide projection.
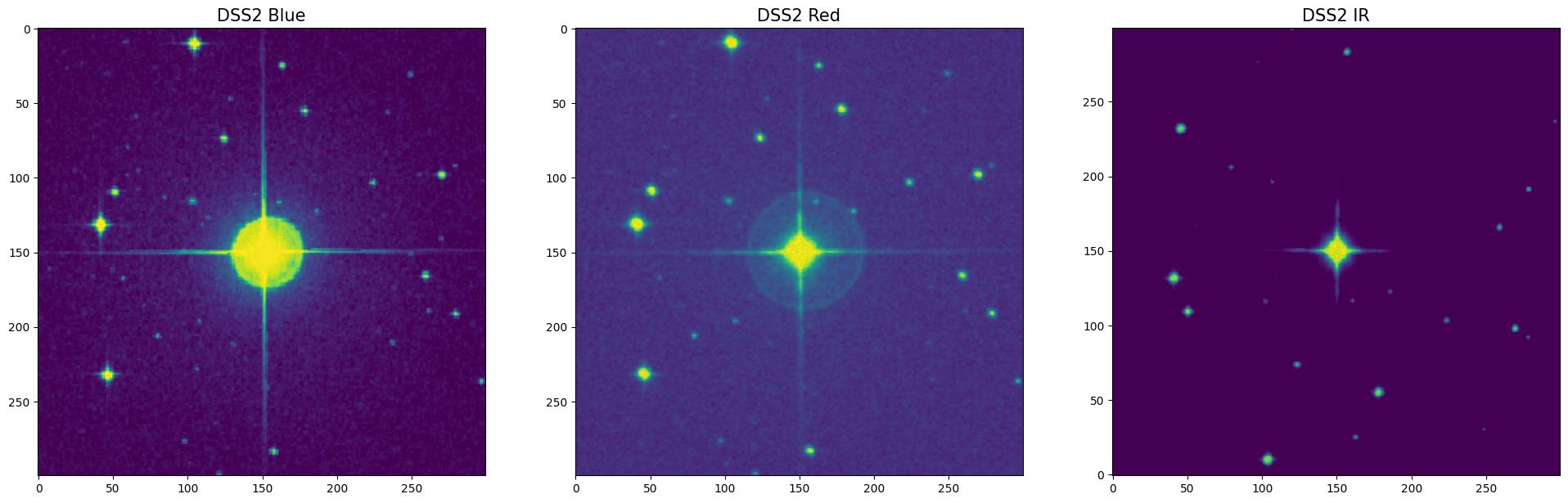
4 Appendix. Check the Field of the Individual Star¶
If you need to visually check an individual star, you can query DSS images manually using the following cells. The default FOV of the image is 6.7’ x 6.7’ (the same as the FOV of the AuxTel).
star_name_img_query = "HD 2527"
FOV=6.7*1/60.0 #6.7 x 6.7 arcminutes for AuxTel
from astroquery.skyview import SkyView
import numpy as np
survey_name = ["DSS2 Blue", "DSS2 Red", "DSS2 IR"]
img = SkyView.get_images(star_name_img_query,survey=survey_name,\
height=FOV*u.degree,width=FOV*u.degree,coordinates='J2000',grid=True,gridlabels=True)
ncol=len(survey_name)
fig,ax = plt.subplots(ncols=ncol,figsize=(24,8))
for i in range(ncol):
plot = ax[i].imshow(img[i][0].data,vmax=np.max(img[i][0].data)*.95,\
vmin=np.max(img[i][0].data)*.25, aspect='equal')
ax[i].set_title(str(survey_name[i]), fontsize=15)
fig.gca().invert_yaxis()
print(star_name_img_query, 'FOV = '+str((FOV*60))+'\"'+'x '+str((FOV*60))+'\"')
References
C. Fabricius, V. V. Makarov, J. Knude, and G. L. Wycoff. Henry Draper catalogue identifications for Tycho-2 stars. \aap , 386:709–710, May 2002. doi:10.1051/0004-6361:20020249.
E. Høg, C. Fabricius, V. V. Makarov, S. Urban, T. Corbin, G. Wycoff, U. Bastian, P. Schwekendiek, and A. Wicenec. The Tycho-2 catalogue of the 2.5 million brightest stars. \aap , 355:L27–L30, March 2000.